DNA Extraction in Oysters for Sequencing Spring 2014
This will be the working journal for the DNA extraction experiments to provide high quality DNA for sequencing.4-10-14
I'll be testing a DNAzol treatment on 5 samples from outplanting in August 2013. Samples have been stored in ethanol at -80C for roughly 9 months.
Protocol is as follows:
Incubation for 24 hours at room temp:
| 1 |
ml |
DNAzol |
| 2.35 |
ul |
Proteinase K (100ug/ml) |
| 1 |
whole |
oyster 5-10 mm |
| X5 |
Reactions |
- Pipette 1 ul DNAzol and 2.35 ul Proteinase K in tube containing oyster
- Homogenize each oyster with disposable pestle.
- Centrifuge briefly
- Incubate overnight at ROOM TEMP.
Isolation:
| 500 |
ul |
Abs EtOH |
| 1 |
ml |
75% EtOH |
| 1 |
ml |
75% EtOH |
| 200 |
ul |
Sterile H2O |
- Centrifuge @ 10,000 g for 10 minutes.
- Collect and Transfer Supernant; Discard pellet
- Add 500 ul Absolute EtOH
- Mix through inversion
- Incubate for 1 minute at ROOM TEMP.
- Spin down DNA precipitate
- Remove supernant
- Wash with 1 ml 75% EtOH
- Spin down
- Remove supernant
- Wash with 1 ml 75% EtOH
- Spin down until solid pellet
- Elute with 200 ul Sterile H2O
- Quant.
Stress Response Testing Fall 2013
This is Jake Heare's Lab Notebook Starting Fall 2013.12-12-13
A Correlation was run between Oyster Size (in mm) with normalized HSP70 concentration.
It was found that Correlation =
| -0.601470235 |
After doing some Stats work in Excel I found that there is a significant difference between the sizes of the sample groups as shown in this ANOVA.
| Anova: Two-Factor With Replication |
||||||
| SUMMARY |
Time 0 |
Time 24 |
Total |
|||
| N |
||||||
| Count |
5 |
5 |
10 |
|||
| Sum |
120 |
130 |
250 |
|||
| Average |
24 |
26 |
25 |
|||
| Variance |
1.5 |
5.5 |
4.222222 |
|||
| S |
||||||
| Count |
5 |
5 |
10 |
|||
| Sum |
83 |
100 |
183 |
|||
| Average |
16.6 |
20 |
18.3 |
|||
| Variance |
2.3 |
4.5 |
6.233333 |
|||
| H |
||||||
| Count |
5 |
5 |
10 |
|||
| Sum |
78 |
81 |
159 |
|||
| Average |
15.6 |
16.2 |
15.9 |
|||
| Variance |
0.3 |
1.7 |
0.988889 |
|||
| Total |
||||||
| Count |
15 |
15 |
||||
| Sum |
281 |
311 |
||||
| Average |
18.73333 |
20.73333 |
||||
| Variance |
16.20952 |
20.78095 |
||||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Sample |
444.8667 |
2 |
222.4333 |
84.46835 |
1.37E-11 |
3.402826 |
|---|---|---|---|---|---|---|
| Columns |
30 |
1 |
30 |
11.39241 |
0.002505 |
4.259677 |
| Interaction |
9.8 |
2 |
4.9 |
1.860759 |
0.177309 |
3.402826 |
| Within |
63.2 |
24 |
2.633333 |
|||
| Total |
547.8667 |
29 |
||||
I've also found that there is a significant difference between the HSP70 Transcript concentration between groups but NOT between Time 0 and Time 24. As seen in this ANOVA.
In the ANOVA below I replaced a value with 0 because it was a huge outlier caused by improper replication due to pipetting error.
| Anova: Two-Factor With Replication |
||||||
| SUMMARY |
Time 0 |
Time 24 |
Total |
|||
| N |
||||||
| Count |
5 |
5 |
10 |
|||
| Sum |
1.929047 |
2.275016 |
4.204063 |
|||
| Average |
0.385809 |
0.455003 |
0.420406 |
|||
| Variance |
0.018329 |
0.034938 |
0.025004 |
|||
| S |
||||||
| Count |
5 |
5 |
10 |
|||
| Sum |
3.87648 |
4.866611 |
8.743092 |
|||
| Average |
0.775296 |
0.973322 |
0.874309 |
|||
| Variance |
0.019266 |
0.095507 |
0.061903 |
|||
| H |
||||||
| Count |
5 |
5 |
10 |
|||
| Sum |
8.144064 |
8.694293 |
16.83836 |
|||
| Average |
1.628813 |
1.738859 |
1.683836 |
|||
| Variance |
0.45257 |
1.297363 |
0.781112 |
|||
| Total |
||||||
| Count |
15 |
15 |
||||
| Sum |
13.94959 |
15.83592 |
||||
| Average |
0.929973 |
1.055728 |
||||
| Variance |
0.428767 |
0.70592 |
||||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Sample |
8.19205 |
2 |
4.096025 |
12.8136 |
0.000164 |
3.402826 |
|---|---|---|---|---|---|---|
| Columns |
0.118608 |
1 |
0.118608 |
0.371041 |
0.54816 |
4.259677 |
| Interaction |
0.021673 |
2 |
0.010836 |
0.033899 |
0.966715 |
3.402826 |
| Within |
7.671893 |
24 |
0.319662 |
|||
| Total |
16.00422 |
29 |
||||
| Anova: Two-Factor With Replication |
||||||
| SUMMARY |
Time 0 |
Time 24 |
Total |
|||
| N |
||||||
| Count |
4 |
4 |
8 |
|||
| Sum |
1.560687 |
1.628999 |
3.189686 |
|||
| Average |
0.390172 |
0.40725 |
0.398711 |
|||
| Variance |
0.024312 |
0.031382 |
0.023952 |
|||
| S |
||||||
| Count |
4 |
4 |
8 |
|||
| Sum |
3.26533 |
3.927122 |
7.192452 |
|||
| Average |
0.816332 |
0.98178 |
0.899056 |
|||
| Variance |
0.014461 |
0.126866 |
0.06839 |
|||
| H |
||||||
| Count |
4 |
4 |
8 |
|||
| Sum |
7.623023 |
8.694293 |
16.31732 |
|||
| Average |
1.905756 |
2.173573 |
2.039665 |
|||
| Variance |
0.092111 |
0.469972 |
0.261386 |
|||
| Total |
||||||
| Count |
12 |
12 |
||||
| Sum |
12.44904 |
14.25041 |
||||
| Average |
1.03742 |
1.187534 |
||||
| Variance |
0.479992 |
0.761679 |
||||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Sample |
11.3175 |
2 |
5.658749 |
44.72711 |
1.04E-07 |
3.554557 |
|---|---|---|---|---|---|---|
| Columns |
0.135206 |
1 |
0.135206 |
1.068678 |
0.314935 |
4.413873 |
| Interaction |
0.063576 |
2 |
0.031788 |
0.251253 |
0.780508 |
3.554557 |
| Within |
2.27731 |
18 |
0.126517 |
|||
| Total |
13.79359 |
23 |
||||
I also ran additional single factor ANOVA's between Time 0 and Time 24 for each population and found no significant difference in HSP70 transcript output.
Northern Pop ANOVA
| Anova: Single Factor |
||||||
| SUMMARY |
||||||
| Groups |
Count |
Sum |
Average |
Variance |
||
| Time 0 |
5 |
1.929047 |
0.385809 |
0.018329 |
||
| Time 24 |
5 |
2.275016 |
0.455003 |
0.034938 |
||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Between Groups |
0.011969 |
1 |
0.011969 |
0.449413 |
0.521492 |
5.317655 |
| Within Groups |
0.213068 |
8 |
0.026634 |
|||
| Total |
0.225038 |
9 |
||||
| Anova: Single Factor |
||||||
| SUMMARY |
||||||
| Groups |
Count |
Sum |
Average |
Variance |
||
| Time 0 |
5 |
3.87648 |
0.775296 |
0.019266 |
||
| Time 24 |
5 |
4.866611 |
0.973322 |
0.095507 |
||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Between Groups |
0.098036 |
1 |
0.098036 |
1.708346 |
0.227516 |
5.317655 |
| Within Groups |
0.459092 |
8 |
0.057386 |
|||
| Total |
0.557128 |
9 |
||||
| Anova: Single Factor |
||||||
| SUMMARY |
||||||
| Groups |
Count |
Sum |
Average |
Variance |
||
| Time 0 |
4 |
7.623023 |
1.905756 |
0.092111 |
||
| Time 24 |
4 |
8.694293 |
2.173573 |
0.469972 |
||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Between Groups |
0.143452 |
1 |
0.143452 |
0.510432 |
0.501789 |
5.987378 |
| Within Groups |
1.686249 |
6 |
0.281041 |
|||
| Total |
1.829701 |
7 |
||||
Additional Population Information
Size and Weight Chart for all Samples
| Sample |
Size (mm) |
Sample weight |
Sample |
Size (mm) |
Sample weight |
| JH_NO_11813_1 |
24 |
0.148 |
JH_N24_11913_1 |
25 |
0.119 |
| JH_NO_11813_2 |
23 |
0.103 |
JH_N24_11913_2 |
26 |
0.147 |
| JH_NO_11813_3 |
24 |
0.105 |
JH_N24_11913_3 |
25 |
0.247 |
| JH_NO_11813_4 |
23 |
0.079 |
JH_N24_11913_4 |
24 |
0.178 |
| JH_NO_11813_5 |
26 |
0.145 |
JH_N24_11913_5 |
30 |
0.218 |
| JH_NO_11813_6 |
23 |
0.125 |
JH_N24_11913_6 |
23 |
0.149 |
| JH_NO_11813_7 |
24 |
0.072 |
JH_N24_11913_7 |
25 |
0.203 |
| JH_NO_11813_8 |
27 |
0.045 |
JH_N24_11913_8 |
21 |
0.158 |
| JH_NO_11813_9 |
28 |
0.138 |
JH_N24_11913_9 |
26 |
0.161 |
| JH_NO_11813_10 |
21 |
0.075 |
JH_N24_11913_10 |
24 |
0.129 |
| JH_NO_11813_11 |
23 |
0.08 |
JH_N24_11913_11 |
25 |
0.251 |
| JH_NO_11813_12 |
30 |
0.186 |
JH_N24_11913_12 |
20 |
0.116 |
| JH_NO_11813_13 |
25 |
0.61 |
JH_N24_11913_13 |
26 |
0.2 |
| JH_NO_11813_14 |
19 |
0.039 |
JH_N24_11913_14 |
25 |
0.248 |
| JH_NO_11813_15 |
21 |
0.101 |
JH_N24_11913_15 |
26 |
0.156 |
| JH_SO_11813_1 |
15 |
0.02 |
JH_S24_11913_1 |
20 |
0.086 |
| JH_SO_11813_2 |
17 |
0.056 |
JH_S24_11913_2 |
18 |
0.128 |
| JH_SO_11813_3 |
15 |
0.05 |
JH_S24_11913_3 |
23 |
0.145 |
| JH_SO_11813_4 |
18 |
0.137 |
JH_S24_11913_4 |
21 |
0.136 |
| JH_SO_11813_5 |
18 |
0.115 |
JH_S24_11913_5 |
18 |
0.102 |
| JH_SO_11813_6 |
19 |
0.17 |
JH_S24_11913_6 |
15 |
0.092 |
| JH_SO_11813_7 |
17 |
0.101 |
JH_S24_11913_7 |
16 |
0.123 |
| JH_SO_11813_8 |
15 |
0.06 |
JH_S24_11913_8 |
19 |
0.128 |
| JH_SO_11813_9 |
16 |
0.154 |
JH_S24_11913_9 |
17 |
0.114 |
| JH_SO_11813_10 |
21 |
0.151 |
JH_S24_11913_10 |
18 |
0.144 |
| JH_SO_11813_11 |
16 |
0.059 |
JH_S24_11913_11 |
19 |
0.068 |
| JH_SO_11813_12 |
15 |
0.015 |
JH_S24_11913_12 |
21 |
0.136 |
| JH_SO_11813_13 |
15 |
0.106 |
JH_S24_11913_13 |
19 |
0.055 |
| JH_SO_11813_14 |
18 |
0.16 |
JH_S24_11913_14 |
17 |
0.138 |
| JH_SO_11813_15 |
14 |
0.07 |
JH_S24_11913_15 |
18 |
0.165 |
| JH_HO_11813_1 |
16 |
0.102 |
JH_H24_11913_1 |
18 |
0.084 |
| JH_HO_11813_2 |
16 |
0.075 |
JH_H24_11913_2 |
17 |
0.074 |
| JH_HO_11813_3 |
15 |
0.084 |
JH_H24_11913_3 |
16 |
0.071 |
| JH_HO_11813_4 |
16 |
0.115 |
JH_H24_11913_4 |
15 |
0.055 |
| JH_HO_11813_5 |
15 |
0.082 |
JH_H24_11913_5 |
15 |
0.068 |
| JH_HO_11813_6 |
19 |
0.087 |
JH_H24_11913_6 |
18 |
0.101 |
| JH_HO_11813_7 |
19 |
0.079 |
JH_H24_11913_7 |
14 |
0.045 |
| JH_HO_11813_8 |
17 |
0.034 |
JH_H24_11913_8 |
17 |
0.088 |
| JH_HO_11813_9 |
16 |
0.058 |
JH_H24_11913_9 |
15 |
0.08 |
| JH_HO_11813_10 |
15 |
0.036 |
JH_H24_11913_10 |
16 |
0.07 |
| JH_HO_11813_11 |
14 |
0.057 |
JH_H24_11913_11 |
16 |
0.057 |
| JH_HO_11813_12 |
15 |
0.062 |
JH_H24_11913_12 |
17 |
0.04 |
| JH_HO_11813_13 |
17 |
0.053 |
JH_H24_11913_13 |
14 |
0.063 |
| JH_HO_11813_14 |
17 |
0.066 |
JH_H24_11913_14 |
15 |
0.028 |
| JH_HO_11813_15 |
15 |
0.075 |
JH_H24_11913_15 |
15 |
0.067 |
12-11-13
Results for the qPCR were as follows:
| Well / Set |
Dye |
Content |
Description |
Efficiency |
C(t) |
Crude Analysis |
Normalized Value |
Normalized Mean Value |
Difference |
St. Dev |
Mean St. Dev |
|
| HSPNO1 |
SBGnew |
Sample |
21.81% |
34.09 |
14.65858 |
0.47612 |
NO |
0.385809 |
0.069194 |
0.121091 |
0.314042 |
|
| HSPNO2 |
SBGnew |
Sample |
33.65% |
30.68 |
156.0213 |
0.225123 |
||||||
| HSPNO3 |
SBGnew |
Sample |
30.74% |
31.12 |
114.9889 |
0.562347 |
||||||
| HSPNO4 |
SBGnew |
Sample |
41.11% |
29.41 |
376.4506 |
0.297098 |
||||||
| HSPNO5 |
SBGnew |
Sample |
36.50% |
30.64 |
160.4102 |
0.36836 |
||||||
| A6 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
#VALUE! |
||||||
| HSPN241 |
SBGnew |
Sample |
30.13% |
30.42 |
186.8512 |
0.40592 |
N24 |
0.455003 |
0.167184 |
|||
| HSPN242 |
SBGnew |
Sample |
39.34% |
27.29 |
1637.767 |
0.582189 |
||||||
| HSPN243 |
SBGnew |
Sample |
25.09% |
31.21 |
108.0309 |
0.47612 |
||||||
| HSPN244 |
SBGnew |
Sample |
16.62% |
31.39 |
95.35248 |
0.164771 |
||||||
| HSPN245 |
SBGnew |
Sample |
34.72% |
27.76 |
1182.192 |
0.646017 |
||||||
| A12 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
#VALUE! |
||||||
| ActNO1 |
SBGnew |
Sample |
32.29% |
33.02 |
30.78761 |
|||||||
| ActNO2 |
SBGnew |
Sample |
55.35% |
28.53 |
693.0491 |
|||||||
| ActNO3 |
SBGnew |
Sample |
48.96% |
30.29 |
204.4805 |
|||||||
| ActNO4 |
SBGnew |
Sample |
56.69% |
27.66 |
1267.092 |
|||||||
| ActNO5 |
SBGnew |
Sample |
34.77% |
29.2 |
435.4718 |
|||||||
| B6 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
|||||||
| ActN241 |
SBGnew |
Sample |
33.90% |
29.12 |
460.3159 |
|||||||
| ActN242 |
SBGnew |
Sample |
54.44% |
26.51 |
2813.118 |
|||||||
| ActN243 |
SBGnew |
Sample |
26.73% |
30.14 |
226.8987 |
|||||||
| ActN244 |
SBGnew |
Sample |
33.89% |
28.79 |
578.698 |
|||||||
| ActN245 |
SBGnew |
Sample |
41.44% |
27.13 |
1829.97 |
|||||||
| B12 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
|||||||
| HSPSO1 |
SBGnew |
Sample |
54.05% |
27.86 |
1102.981 |
0.784477 |
SO |
0.775296 |
0.198026 |
0.124147 |
||
| HSPSO2 |
SBGnew |
Sample |
55.69% |
26.63 |
2588.475 |
0.913785 |
||||||
| HSPSO3 |
SBGnew |
Sample |
65.65% |
25.88 |
4354.557 |
0.907469 |
||||||
| HSPSO4 |
SBGnew |
Sample |
49.31% |
29.2 |
435.4718 |
0.659599 |
||||||
| HSPSO5 |
SBGnew |
Sample |
56.02% |
27.49 |
1425.647 |
0.61115 |
||||||
| D6 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
#VALUE! |
||||||
| HSPS241 |
SBGnew |
Sample |
57.88% |
26.8 |
2300.594 |
1.37579 |
S24 |
0.973322 |
0.276416 |
|||
| HSPS242 |
SBGnew |
Sample |
58.54% |
26.12 |
3686.855 |
1.189323 |
||||||
| HSPS243 |
SBGnew |
Sample |
53.98% |
27.81 |
1141.9 |
0.65504 |
||||||
| HSPS244 |
SBGnew |
Sample |
61.01% |
27.9 |
1072.803 |
0.706968 |
||||||
| HSPS245 |
SBGnew |
Sample |
51.20% |
26.94 |
2087.719 |
0.93949 |
||||||
| D12 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
#VALUE! |
||||||
| ActSO1 |
SBGnew |
Sample |
63.47% |
27.51 |
1406.009 |
|||||||
| ActSO2 |
SBGnew |
Sample |
66.04% |
26.5 |
2832.696 |
|||||||
| ActSO3 |
SBGnew |
Sample |
73.57% |
25.74 |
4798.572 |
|||||||
| ActSO4 |
SBGnew |
Sample |
50.38% |
28.6 |
660.2068 |
|||||||
| ActSO5 |
SBGnew |
Sample |
72.13% |
26.78 |
2332.727 |
|||||||
| E6 |
SBGnew |
Sample |
25.05% |
39.39 |
0.371306 |
|||||||
| ActS241 |
SBGnew |
Sample |
46.98% |
27.26 |
1672.199 |
|||||||
| ActS242 |
SBGnew |
Sample |
61.53% |
26.37 |
3099.959 |
|||||||
| ActS243 |
SBGnew |
Sample |
49.74% |
27.2 |
1743.251 |
|||||||
| ActS244 |
SBGnew |
Sample |
57.93% |
27.4 |
1517.47 |
|||||||
| ActS245 |
SBGnew |
Sample |
60.46% |
26.85 |
2222.184 |
|||||||
| E12 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
|||||||
| HSPHO1 |
SBGnew |
Sample |
41.80% |
29.72 |
303.6239 |
2.204794 |
HO |
1.628813 |
0.54476 |
0.601711 |
||
| HSPHO2 |
SBGnew |
Sample |
50.26% |
27.74 |
1198.704 |
1.484855 |
||||||
| HSPHO3 |
SBGnew |
Sample |
65.62% |
25.57 |
5399.033 |
1.932591 |
||||||
| HSPHO4 |
SBGnew |
Sample |
69.68% |
25.9 |
4294.573 |
2.000783 |
||||||
| HSPHO5 |
SBGnew |
Sample |
47.00% |
27.4 |
1517.47 |
0.521041 |
||||||
| G6 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
#VALUE! |
||||||
| HSPH241 |
SBGnew |
Sample |
46.23% |
28.01 |
994.0037 |
2.071381 |
H24 |
2.173573 |
0.5937 |
|||
| HSPH242 |
SBGnew |
Sample |
48.31% |
27.17 |
1779.902 |
2.909699 |
||||||
| HSPH243 |
SBGnew |
Sample |
57.73% |
26.62 |
2606.49 |
2.429607 |
||||||
| HSPH244 |
SBGnew |
Sample |
53.12% |
26.68 |
2500.253 |
1.283607 |
||||||
| HSPH245 |
SBGnew |
Sample |
44.38% |
26.73 |
2415.038 |
18.15583 |
||||||
| G12 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
#VALUE! |
||||||
| ActHO1 |
SBGnew |
Sample |
35.51% |
30.86 |
137.7108 |
|||||||
| ActHO2 |
SBGnew |
Sample |
34.65% |
28.31 |
807.2871 |
|||||||
| ActHO3 |
SBGnew |
Sample |
49.30% |
26.52 |
2793.676 |
|||||||
| ActHO4 |
SBGnew |
Sample |
62.15% |
26.9 |
2146.446 |
|||||||
| ActHO5 |
SBGnew |
Sample |
50.58% |
26.46 |
2912.38 |
|||||||
| H6 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
|||||||
| ActH241 |
SBGnew |
Sample |
35.94% |
29.06 |
479.8749 |
|||||||
| ActH242 |
SBGnew |
Sample |
36.25% |
28.71 |
611.7134 |
|||||||
| ActH243 |
SBGnew |
Sample |
40.92% |
27.9 |
1072.803 |
|||||||
| ActH244 |
SBGnew |
Sample |
43.94% |
27.04 |
1947.834 |
|||||||
| ActH245 |
SBGnew |
Sample |
18.54% |
30.91 |
133.0173 |
|||||||
| H12 |
SBGnew |
Sample |
N/A |
N/A |
#VALUE! |
|||||||
The qPCR ran fine although the results are a little odd. I did have one contamination event in a negative control but it did not appear in the second negative control for that group. I also had a pipetting error on the last wells the Hood Canal Population and the associated negative control. This is reflected in the normalized value for H245 as it was nearly 9 fold larger than ever single other sample in the set. Time point 24 for the Hood Canal samples only have 4 replicates instead of five because of the outlier caused by pipetting error.
It appears that the Northern animals had much lower concentrations of Actin than HSP70, Southern animals had a close to 1:1 ratio of HSP to Actin, and Hood Canal Animals had the highest amount of HSP to Actin. You can see in the graph below that all populations showed an increasing value of HSP70 from Time 0 to Time 24.
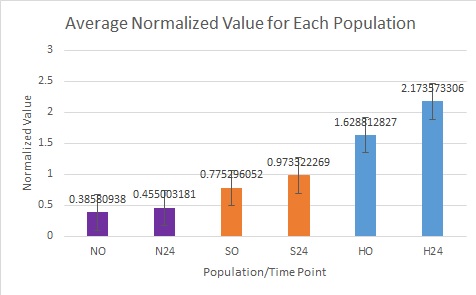
The smallest difference was seen in the Northern animals, with the largest difference being found in the Hood Canal animals.

From this data, It seems that Hood Canal (Dabob) oysters have a stronger reaction to Heat Shock than the Northern or Southern populations. This could be due to the isolated nature of Dabob Bay since it is not directly influenced by currents coming off the sound or the Strait of Juan de Fuca.
12-10-2013
38 reactions per primer set (2 primers, 30 samples each, 2 control for each population, 2 extra reaction for pipette error)
Ssofast Evagreen MM 10 ul X38 380 ul
Fwd Primer 0.5 ul X38 19 ul
Reverse Primer 0.5 ul X38 19 ul
Nuclease Free H20 8 ul X38 304 ul
cDNA 1 ul
I used the program Sybr New Plate + Sybr cDNA 55 melt 2 reads on the opticon.
11-29-13
Findings from the qPCR.
| Well / Set |
Dye |
Content |
Description |
Efficiency |
C(t) |
Crude Analysis |
Normalized Info |
Fold Difference |
Difference between 0 and 24 |
Percent Increase |
||
| HSP70_N0_1 |
SBGnew |
Sample |
54.33% |
30.05 |
241.5127 |
2.000783 |
4.115737 |
1.000783 |
||||
| HSP70_N24_1 |
SBGnew |
Sample |
63.03% |
27.19 |
1755.383 |
0.574169 |
1.181102 |
-1.42661 |
-0.42583 |
1513.871 |
626.8286 |
|
| HSP70_S0_1 |
SBGnew |
Sample |
66.46% |
27.23 |
1707.356 |
0.48613 |
0.999999 |
-0.51387 |
||||
| HSP70_S24_1 |
SBGnew |
Sample |
71.05% |
25.39 |
6116.909 |
0.763013 |
1.569566 |
0.276884 |
-0.23699 |
4409.553 |
258.268 |
|
| HSP70_H0_1 |
SBGnew |
Sample |
62.79% |
28.46 |
727.5251 |
0.562347 |
1.156782 |
-0.43765 |
||||
| HSP70_H24_1 |
SBGnew |
Sample |
65.31% |
27.56 |
1358.088 |
0.619686 |
1.274734 |
0.05734 |
-0.38031 |
630.5631 |
86.67235 |
|
| Actin_NO_1 |
SBGnew |
Sample |
58.19% |
31.05 |
120.7091 |
|||||||
| Actin_N24_1 |
SBGnew |
Sample |
89.68% |
26.39 |
3057.257 |
2936.548 |
2432.748 |
|||||
| Actin_SO_1 |
SBGnew |
Sample |
93.06% |
26.19 |
3512.141 |
|||||||
| Actin_S24_1 |
SBGnew |
Sample |
118.80% |
25 |
8016.781 |
4504.639 |
128.259 |
|||||
| Actin_HO_1 |
SBGnew |
Sample |
90.21% |
27.63 |
1293.731 |
|||||||
| Actin_H24_1 |
SBGnew |
Sample |
82.39% |
26.87 |
2191.573 |
897.8423 |
69.39945 |
|||||

I have a PPower point presentation explaining in some detail the findings of the qPCR
Oly Heat Shock Presentation
11-25-13
qPCR test on Thursday results were analyzed this morning. It looks like the primers work with the reagents just fine though amplication did not occur in samples tested until the mid to late 20 cycles range. I'm going to run a final qPCR on my samples this afternoon using the same protocol as follows:
15 reactions (2 primers, 6 samples each, 1 control each, 1 extra reaction for pipette error)
Ssofast Evagreen MM 10 ul X15 150 ul
Fwd Primer 0.5 ul X15 7.5 ul
Reverse Primer 0.5 ul X15 7.5 ul
Nuclease Free H20 8 ul X15 120 ul
cDNA 1 ul
I used the program Sybr New Plate + Sybr cDNA 55 melt 2 reads on the opticon.
11-21-13
Collected primers from Mackenzie and performed PCR on six samples for all 4 primers.
All reactions were run on a Agarose gel using the following protocol.
- Measure out 7 ml of 25X TAE
- Add to 250 ml beaker and raise volume to 175 ml with nanopure water.
- Add 2 g standard agarose
- Microwave for 2:30 seconds, swirl 30 seconds, microwave another 30 seconds.
- Allow agarose to cool slightly and then add 17.5 ul EtBr to solution. Swirl 30 seconds.
- Quickly and evenly poor gel into casting tray with 2 rows of combs (I used one 20 well comb and one 16 well comb). Remove bubbles if necessary
- Allow gel to cool to room temp.
- Place gel in electrophoresis box and fill box with 750 ml 1X TAE until the gel is full covered.
- Remove combs.
- Pipette in 5 ul Ladders and 12.5 ul samples
- Run gel at 100v for 40 mins.
- Illuminate bands on transilluminator in darkened room. Take pictures.
Gels were set up as follows
Glycogen Phosphorylase Glutathione Peroxidase
Neg Lad N01 N241 S01 S241 H01 H241 Neg Lad N01 N241 S01 S241 H01 H241
HSP70 Actin
Neg Lad N01 N241 S01 S241 H01 H241 Neg Lad N01 N241 S01 S241 H01 H241
After running the samples I found that:
Glycogen phosphorylase had double bands in all the samples as well as a indiscriminate replication smear in the low weight area.
Glutathione peroxidase had an indiscriminate replication smear in the low weight region.
HSP70 had strong low weight bands indicative of target appropriate replication.
Actin had strong low weight bands indicative of target appropriate replication.
It appears that Glycogen Phosphorylase and Glutathione peroxidase do not work with my samples while HSP70 and Actin do.
After these findings I set up a very limited qPCR reaction with only 4 reactions occuring. 2 with HSP70 and 2 with Actin, each having a positive control sample and a negative control water. I ran them using the following protocol:
Ssofast Evagreen MM 10 ul
Fwd Primer 0.5 ul
Reverse Primer 0.5 ul
Nuclease Free H20 8 ul
cDNA 1 ul
I used the program Sybr New Plate + Sybr cDNA 55 melt 2 reads on the opticon.
11-15-13
cDNA Quantification and PCR
cDNA will be quantified on the nanodrop today. It has been stored at -80 since tuesday.
After quantification has occurred. I will keep the first cDNA sample from each population at each time point for use in PCR and the rest will go back to the -80.
(Six samples total will be used for PCR, with one negative control)
Primers will be rehydrated using the following procedure:
- Find nMole concentration on data sheet for each primer stock.
- Multiply nM number by 10 and add that many ul of Nuclease Free Water to the primer.
- Vortex
- Sample should now be at 1 uM per uL.
- Take 10 ul SS and add it to 90 ul Nuclease Free H2O.
- This will make a working stock with a 0.1 uM per uL concentration.
- Put SS into -20. Keep WS out for PCR.
PCR will go as follows
Master Mix for 1 reaction (There will be 6 samples and 1 neg control per individual primer PCR so make enough for 8 reactions)
- 12.5 ul goTaq Green
- 1 ul WS Primer F
- 1 ul WS Primer R
Add 14.5 ul MASTER Mix to tube
Add <250 ng cDNA (volume added based on concentration of cDNA)
Add supplementary water to create 25 ul reaction mixture.
Run the following program (To Be Named Later):
2 min 95 C Initial Denaturation
45 sec 95 C
45 sec 54 C Repeat these steps 30 times
2 min 73 C
5 min 74 C Final Extension
FRVR 4C Holding temp until samples can be retrieved from thermocycler.
11-12-13
cDNA was created by the following protocol.
- In 0.5 ml tubes add 4 ul nuclease free water and 1 ul oligo DT.
- Then add 5 ul RNA.
- Incubate 5 min at 70C then immediately transfer to ice.
- Create a master mix of the following
- 175 ul MMLV 5x Reaction Buffer
- 175 ul dNTP
- 35 ul MMLV Reverse Transcriptase
- 140 ul Nuclease/RNA Free H2O
- Aliquot 15 ul of MM to each sample tube.
- Incubate at 42 C for 60 min. Then increase to 70 C for 3 min.
- Spin Down.
- Store.
11-11-13
Samples one 1-5 from each population in each time point were thawed and heat block turned to 55 C.
During the thawing process, UV hood and work materials were sterilized using the following procedure:
- Hood work surface wiped with 75% EtOH and RNase Zap (wiping from center to edges in a spiral out pattern)
- All tools, pipettes, tubes, tube racks, waste containers, etc (anything going under the hood) wiped with EtOH and RNase Zap.
- All items placed under hood, blower turned on, and then the UV light turned on.
- UV light on for 15 minutes to sterilize equipment.
As UV sterilized hood, Thawed Samples had 500 ul Trizol added to them and then vortexed.
After UV sterilization completed. Samples were placed under hood and the RNA isolation protocol was followed as such.
- 200 ul Chloroform was added to each tube. (Under hood)
- Tubes vortexed for 30 seconds.
- Incubate tubes for 5 minutes at room temp.
- Centrifuge 15 mins at 16.1 rcf and 4C. (15 samples at a time in centrifuge, other 15 sat in fridge or moved to next step)
- Extract clear top portion and place in new RNase Free tube. (Roughly 500 ul) (Under hood)
- Add 500 ul Isopropanol to each tube (Under hood)
- Vortex 15 seconds.
- Incubate tubes 10 mins at room temp.
- Spin for 8 minutes at 16.1 rcf and 4C
- Remove supernatant waste and dispose. (Under hood)
- Add 1 ml 75% EtOH. (Under hood)
- Vortex 30 seconds
- Centrifuge at 7.5 rcf for 5 mins at 4C
- Remove supernatant waste and dispose. (Under Hood)
- Spin for 30 seconds at room temp to pool excess EtOH.
- Draw off excess EtOH with 100 ul pipette. (Under hood)
- Allow to air dry under hood for 5 minutes to evaporate remaining EtOH. (Under Hood)
- Resuspend in 100 ul 0.1% DEPC H2O. (Under hood)
- Incubate at 55C for 5 minutes.
- Vortex
- Store in -80 C for later use.
RNA was stored in -80 C for later use.
11-9-2013
Samples were taken at roughly the 24 hour mark after the heater was switched on in the tank. No visible mortalities in the remaining animals.
Animals were then measured for length and dissected using snips, scissors, spatula, and forceps under sterile conditions. Whole body tissue samples were then placed in labelled tubes and flash frozen on dry ice. Samples were weighed to determine sample weight taken.
Labeling is as follows:
North Sound
JHN24119131-15
South Sound
JHN24119131-15
Hood Canal
JHN24119131-15
The first 5 samples of each group were thawed and mixed with 500 ul Trizol. Homogenized for 30 seconds to one minute and placed on ice.
All samples were then stored in -80 C freezer for later use.
11-8-2013
Animals have been fed and conditioned at 12 C for the last 7 days and fed once every 24 hours.
At 3 pm time point 0 animals were collected and the heating element was turned on to 23 C and fed one last time.
Time point animals were measured for length and dissected using snips, scissors, spatula, and forceps under sterile conditions. Placed in labelled tubes and placed on dry ice for flash freezing. Samples were then weighed on a scale to determine weight of the tissue taken.
Labeling system goes as follows:
North Sound
JHN0118131-15
South Sound
JHS0118131-15
Hood Canal
JHH0118131-15
Samples were flash frozen on dry ice. Samples 1-5 were thawed, placed in 500 ul of Trizol, homogenized for 1.5 minutes until well homogenized.
All samples stored at -80 C.
11-5-2013
Animals in tank look good though there is a small amount of proteinaceous material in the water causing foaming around the bubblers. They have been fed once roughly every 24 hours.
I have received my primers today for the three experimental sequences. They are in the -20 C fridge in FTR 209 in a box labelled Jake's PCR Primers and Other Goodies.
Claire has notified me the primers need to be rehydrated using the following steps.
- Find nanomolar concentration on data sheet for associated primer. (some primer names are missing the forward/reverse mark in the name. Use the sequence data to match data sheet to primer)
- Using the specified concentration (ie. 35.2 nM) multiply by ten (ie. 35.2 X 10 = 352) and this equals the amount of RNase/DNase Free Water needed rehydrate the primer into stock solution.
- Before adding water, centrifuge down concentrated primer.
- Add appropriate amount of water to primer stock. This is the STOCK SOLUTION (SS)
- To create working stock, dilute SS in a 1 in 10 dilution (10 ul SS in 90 ul H20). This is the WORKING STOCK (WS).
- WS is used for qPCR and PCR.
11-1-2013
I travelled to Port Gamble on Friday november 1st. I collected 30 samples per population of small seed oysters. Labelling and Sizes are as follows:
South Sound: Orange Ziptie, 15-25 mm in size. Tag ID: SN
North Sound: Purple/Burgundy Ziptie, 30-45 mm in size. Tag ID: NF2
Hood Canal: Blue Ziptie, 10-20 mm in size. Tag ID: H13-0607 (Jun 7th)
Acclimation set up:
Large rubbermaid tub with 10-15 gallons of sea water at 53F with 2 aerators at constant flow rate. Bagged animals scattered randomly through tub.
Animals are fed once every 24 hours or so with commercial shellfish diet. They have been at these conditions since 6 pm Friday and will stay this way until experiment can be performed.
Experiment design.
At time 0, 15 animals from each population will be sacrificed and whole body tissues or 1 gram (whichever is less) will be placed in labelled tubes and frozen for later extraction. Remaining animals will have the temperature in the tank raised 10 C and will be left at that for 24 hours. Then the remaining animals will be sampled and stored for later extraction.
10-29-2013
This week I designed primers for Glycogen phosphorylase based on the protein sequence for it in C. gigas. I designed a primer for HSP70 in O. lurida based on the protein sequence for it in O. edulis. I designed a primer for Glutathione S Transferase sigma based on its nucleotide sequence in C. gigas.
Friday I will be traveling to Port Gamble to pick up animals from the three populations (roughly 60) and bringing them back to the lab to acclimate in the 12 C cold room until next week.
10-21-13
Page Gels and Western Blots
I was unable to attend this weeks lab due to field work. I have done extensive work with PAGE gels and western blots in the past.
10-15-13
qPCR and Protein Extraction
Lab Summary
We covered qPCR theory and techniques, protein extraction, and began planning for our semester project.
Materials
- Pipettes
- Filter Tips
- 0.5 ml PCR white tube strips w/ clear lid strips
- cDNA Sample
- 1.5 ml RNase Free Tubes
- Nuclease Free H2O
- Thermocycler
- 2X Ssofast EvaGreen Master Mix
- pestles
- lysis buffer
- Forward Primer for DNMT
- Reverse Primer for DNMT
Methods
qPCR
Prepare master mix for 5 reactions. Single reaction volumes as listed.
- 12.5 ul SsoFast EvaGreen SupaMix
- 0.5 ul Foward Primer
- 0.5 ul Reverse Primer
- 10.5 ul H2O
- Add master mix to 4 tubes in strip.
- Add 1 ul cDNA to first two tubes
- Add 1 ul H2O for two negative control tubes
- Secure and clean cap
- Spin down
- Load on Plate
- Run Program
- 95 C 10 mins
- (95 C 15 sec, 55 C 15 sec, 72 C 15 sec) 39 times
- 95 C 10 sec
- Melt curve (65 to 95 C at 0.5 C per 5 sec)
Protein Extraction
- Collect tissue samples and record weight
- Add 500 ul Lysis Buffer
- Homogenize
- Invert tube
- Spin for 10 min at Max Speed at 4C
- Transfer Supernatant to fresh tube
- Label tube, store at -20 C.
For My Experimental Design
4 Tanks2 Heaters
1 canister of nitrogen
Tubing with Dividers to bubble in Nitrogen
Olympia Oysters (40)
Aerators for the other two tanks
Timer
Control group will be in one tank at ambient temperature with aeration
Hypoxia group will be at ambient temp with DO reduced to 3-4% by Nitrogen bubbling
Heat Group will be at 30 C with aeration
Combined will be at 30 C with DO at 3-4%
Samples taken at 0, 6, and 12 hour mark. qPCR for HIF mRNA's in Olys. Possible protein analysis for HIFs in situ.
10-8-13
RNA Extraction and cDNA Creation
Lab Summary
In this lab we complete the RNA extraction that we started last week as well as create complementary DNA strands of the RNA for use in PCR.
Materials
Pipettes
Filter Tips
1.5 ml Tubes
0.5 ml Tubes
Tube racks
Ice Bucket
Chloroform
Hot Water Bath
Thermocycler
RNase Free Water
isopropanol
75% EtOH
0.1% DEPC H2O
Oligo dT
MMLV-RT
Buffer
NTPs
Methods
RNA Extraction
- Incubate previous weeks trizol mixture until thawed
- add 200 ul chloroform
- vortex till milky
- incubate at RT for 5 mins
- Spin 14,000 G at 4C for 15 mins
- Transfer clear aqueous layer to second 1.5 ml tube.
- Add 500 ul isopropanol
- inversion mixing
- Incubate at RT for 10 min
- Spin for 8 mins, 14,000 G at 4C
- Remove supernatant
- Add 1ml 75% EtOH to pellet. Vortex.
- Spin 7500 G for 5 mins at 4C
- Remove EtOH supernatant.
- Spin for 15 s and remove remaining EtOH supernatant.
- Leave tube open for 4 mins to off gas EtOH.
- Resuspend in 100 ul 0.1% DEPC H2O
- Incubate at 55 C for 5 mins
- Flick mix and store on Ice
cDNA Creation
- Make a solution of 5 ul RNA, 1 ul oligo dT, 4 ul RNase Free H2O.
- Incubate solution at 70 C for 5 mins.
- Add 14 ul Master Mix (5 ul buffer, 5 ul NTPs, 0.5 ul MMLV-RT, 3.5 ul H2O) to RNA solution.
- Incubate at 42 C for 60 mins. Then increase to 70 C for 3 mins
- Spin down
- Store at -20
Conclusions.
It was relatively easy to extract the RNA as well as create the cDNAs. I'm not sure about the RNA quality/quantity as I skipped the quantifying on the Nanodrop. I will quantify for the actual project.
For my project I'm going to look at the effects of hypoxia and heat stress on the upregulation of RNA transcripts for Hypoxia Induced Factors in Olympia Oysters. My null hypothesis is that having additional heat stress will not change the regulation of HIFs noticeably under hypoxic conditions. My alternative hypothesis is that heat stress will cause an increase in HIFs during hypoxia. My second alt hypothesis is that heat stress will cause a denaturing of HIFs and actually cause a reduction in HIFs under hypoxic conditions.
10-1-13
DNA/RNA Extraction Lab
Lab Summary
This is the first lab for Integrative Environmental Physiology. In this lab we are reviewing DNA and RNA extraction techniques using DNazol and Trizol (phenol/chloroform) extractions. Extraction products will then be quantified and purity will be assessed using a Nanodrop spectrophotometer.
Materials
Pipettes (1000ul, 100ul, 10ul)
Sterile RNase-Free pipette tips
Sterile 1.5 ml sample tubes
Sterile disposable pestles
Vortexer
Microcentrifuge
Ice Bucket
Sterile Gloves
Lab Pen
Safety Glasses
Trizol
DNazol
Absolute Ethanol
75% Ethanol
0.1% DEPC H2O
Kimwipes
Nanodrop
Methods
RNA Extraction (Part 1, Part 2 takes place in Week 2 lab)
1. Label tube containing 80 mg olympia gill tissue with RNA, species, tissue type, date, my initials, and an L for lab.
2. Add 500 ul Trizol to tissue.
3. Homogenize thoroughly and vortex. Put on ice.
4. Add 500 ul Trizol and store at -80C until next week.
DNA Extraction
- Tissue tube contains 77 mg olympia gill tissue. Label all tubes with DNA, species, tissue type, date, my initials, L for lab.
- Add 500 ul DNazol to tissue.
- Homogenize.
- Add 500 ul DNazol to homogenate.
- Incubate at room temp for 5 mins.
- Spin at 10,000 G at room temp for 10 mins.
- Transfer supernatant to clean tube.
- Add 500 ul absolute EtOH.
- Mix through inversion.
- Incubate 1 min. at room temp.
- Remove DNA using pipette and place in new tube.
- Add 1 ml 75% EtOH to tube and mix through inversion.
- Pipette off lysate.
- Add 1 ml 75% EtOH to tube and mix through inversion.
- Centrifuge for 1 min.
- Pipette off supernatant.
- Resuspend pellet in 300 ul 0.1% DEPC H2O through inversion and vortexing.
- Quantify using Nanodrop.
DNA Quantification
- Clean pedestal with kimwipe
- Add 2 ul 0.1% DEPC water to pedestal. Initiate Nanodrop.
- Wipe off pedestal.
- Add 2 ul 0.1% DEPC water to pedestal. Measure BLANK.
- Wipe off pedestal.
- Add 2 ul of sample.
- Measure.
- Record A260/A280, A260/A230, and concentration.
- Place tube in rack to be stored in -20C freezer for later use.
Findings.
RNA is being saved for next week.
DNA Quantification.
Concentration: 280 ng/ul
A260/A280 Ratio: 1.84
A260/A230 Ratio: 0.79
I had high concentration of DNA in my extraction. It was also relatively clean at a A260/A280 ratio well within the 1.7-1.9 range. I feel prepared to complete the RNA extraction next.
Additional Material.
Prior to next weeks lab, we have been tasked with determining a few genes we would like to create primers for and explaining why they are important.
I wish to look at the HSP70 and HSP90 genes for heat shock proteins. These proteins help maintain proper folding of vital proteins within the cells. They also contain Heat Shock Factors which help target portions of the genome to be expressed during times of sublethal heat stress.
I also would like to look at Catalase coding genes as a way to measure general stress on the animals from any number of stressors.
Finally I'd like to look at Hypoxia Induced Protein coding genes to determine the reaction Olympia oysters have toward low DO contents in shallow tidal areas.